Hybrid Codes
Python is a general-purpose programming language not specifically targeted to particular applications in scientific and numerical computing. The ability to incorporate compiled extension modules into a Python program, however, provides many opportunities to improve numerical performance, by allowing for more code to run in a compiled extension module rather than through interpreted bytecodes. The resulting codes are hybrids that straddle interpreted and compiled code layers, coupled through the Python/C API. The overall performance of such a code depends on the granularity of computations in compiled code and the overhead required to communicate between languages.
The developer and user communities have created a rich ecosystem of third-party libraries for numerical and scientific computing, data analysis, machine learning, and data visualization. While some of that functionality is written in Python, many of the core number-crunching parts are extension modules written in C or Fortran. Furthermore, by structuring function calls so that many operations are executed within a compiled function before returning to the Python interpreter, numerical efficiencies can be achieved, often on a par with code written completely in a compiled language. Conversely, if only a few operations are performed by the compiled code, then the overhead of making the call might negate any performance advantage.
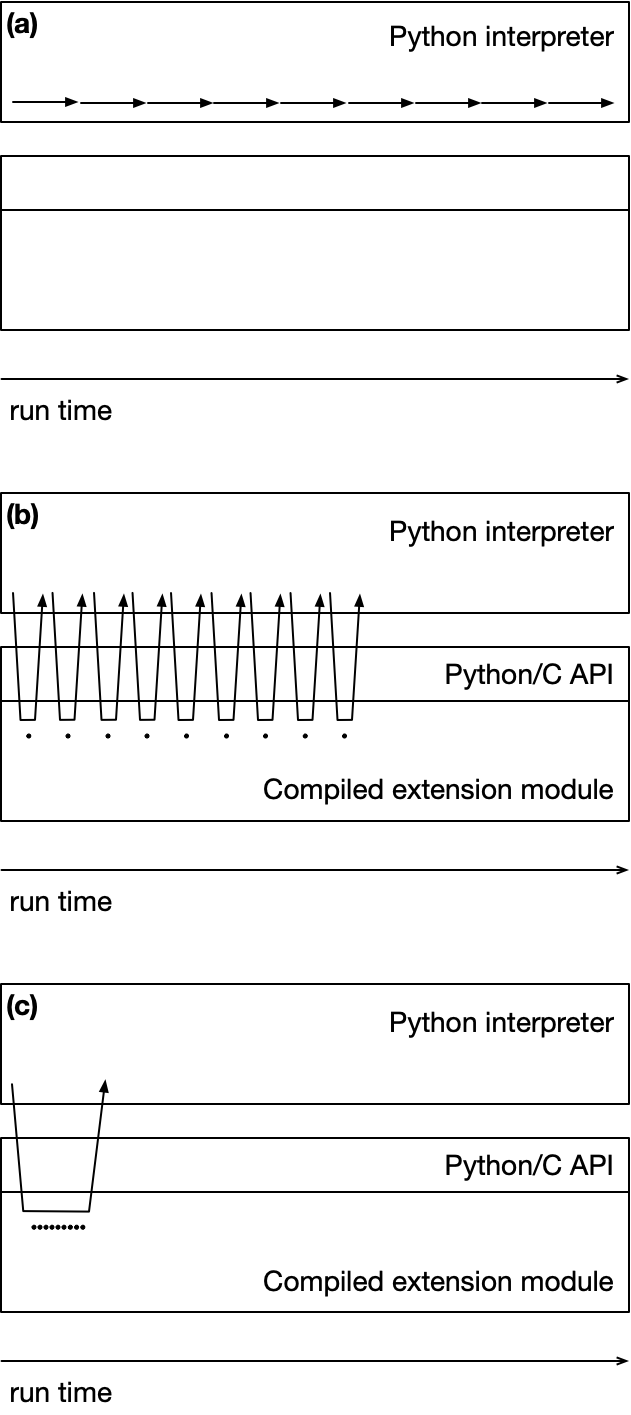
This is illustrated schematically in the figure below, representing calls from the Python interpreter, through the Python/C API, into a compiled extension module (e.g., written in C, or some other compiled language which can be called from C). The three panels are intended to convey three different scenarios where the same computation is performed, but through a different set of function calls, which require different amounts of run time to complete. Panel (a) at the top represents a "pure Python" implementation; panel (b) in the center represents a scenario where multiple calls to compiled functions are being made with results returned to Python; and panel (c) at the bottom represents a situation where all those compiled function calls are "bundled" together and triggered by a single function call from Python. For the sake of concreteness here, imagine that we are looping through a container with 9 elements and performing some computation on those elements. In panel (a), these computations in pure Python are relatively slow and take a longer time to complete. In panels (b) and (c), these computations are carried out in the compiled extension module, and are fast (represented by small black dots); in (b) and (c), the arrows represent data movement from the interpreter to the compiled layer and back again. In panel (b), each separate computation is triggered by a separate function call from the interpreter; for example, we might be executing a for loop in Python and accessing each element one-at-a-time in that loop. In panel (c), there is a single call from Python that triggers all 9 computations before returning all the data back to Python. Because of the overhead associated with converting from Python to C and back for each function call, the run time for the computation in panel (b) will take longer than that in panel (c), where the translation overhead is spread out or amortized over all 9 computations. As noted, the speedup gained depends in part on the granularity of computations in compiled code and the overhead required to communicate between the layers.
CVW material development is supported by NSF OAC awards 1854828, 2321040, 2323116 (UT Austin) and 2005506 (Indiana University)