Reading Arrays and Tables
I/O for NumPy Arrays
If you are working just with NumPy arrays, there are a variety of functions for array I/O, described in the NumPy documentation. These functions differ in whether data are stored in a file in plain text, a binary .npy file, a compressed .npz file, or in a text file with missing data.
I/O for Arrays in Other Formats (including HDF5)
Array data, or bundled and annotated collections of arrays, are sometimes produced and/or stored in other formats, such as HDF5, MATLAB .mat, IDL, and NetCDF. The h5py module provides a Python interface to HDF5 files, extracting named data sets into numpy arrays. The scipy.io module provides functions for reading and writing MATLAB style mat files (version 4 through 7.1), IDL files, Matrix Market files, unformatted Fortran files, netcdf files, Harwell-Boeing files, WAV sound files, and arff files. It should be noted that Version 7.3 MATLAB MAT-files use an HDF5 based format, and the h5py module allows you to read data in that format into Python.
I/O for Pandas Dataframes
To deal with tabular data, Pandas provides a number of useful functions for reading data in different formats, including (but not limited to):
read_csv- read a CSV (comma-separated) flat file into a DataFrame
read_excel- read an Excel table into a DataFrame
read_sql_query- read the results of a SQL query on a database into a DataFrame
In addition, there are corresponding output methods for writing dataframes to files in various formats (e.g., DataFrame.to_csv(), DataFrame.to_excel(). Online documentation provides more information on the available Pandas I/O functions.
In this page and the next, we will examine some of these input functions with our different datasets.
Reading CSV-Formatted Data Into Dataframes
Because the Baseball Databank data are all stored in csv files, we can make use of the pandas read_csv function to ingest those data. As noted, the data are not just in one csv file, but 27 of them. Fortunately, the glob module — part of the Python Standard Library — provides a convenient function that can be used to identify all csv files in a specified directory, without our having to type their names in ourselves. The following code defines and then calls a function that uses glob.glob in order to get the names of all the csv files in a specified directory, and then the pd.read_csv function to convert each file to a pandas DataFrame. All the DataFrames are stored and returned in a Python dictionary named dfs, keyed on the name of the associated csv file (stripped of the trailing ".csv" suffix):
In order to facilitate access to a few dataframes that we want to work with, we can create a few variables to refer to those tables more easily:
# extract a few for further processing
batting = bbdfs['Batting']
pitching = bbdfs['Pitching']
teams = bbdfs['Teams']
When working with a new dataset, it's usually good to poke around to get a feel for what the data contains. We can peek at the first few rows of some of these with the head() method (default number of rows is 5):
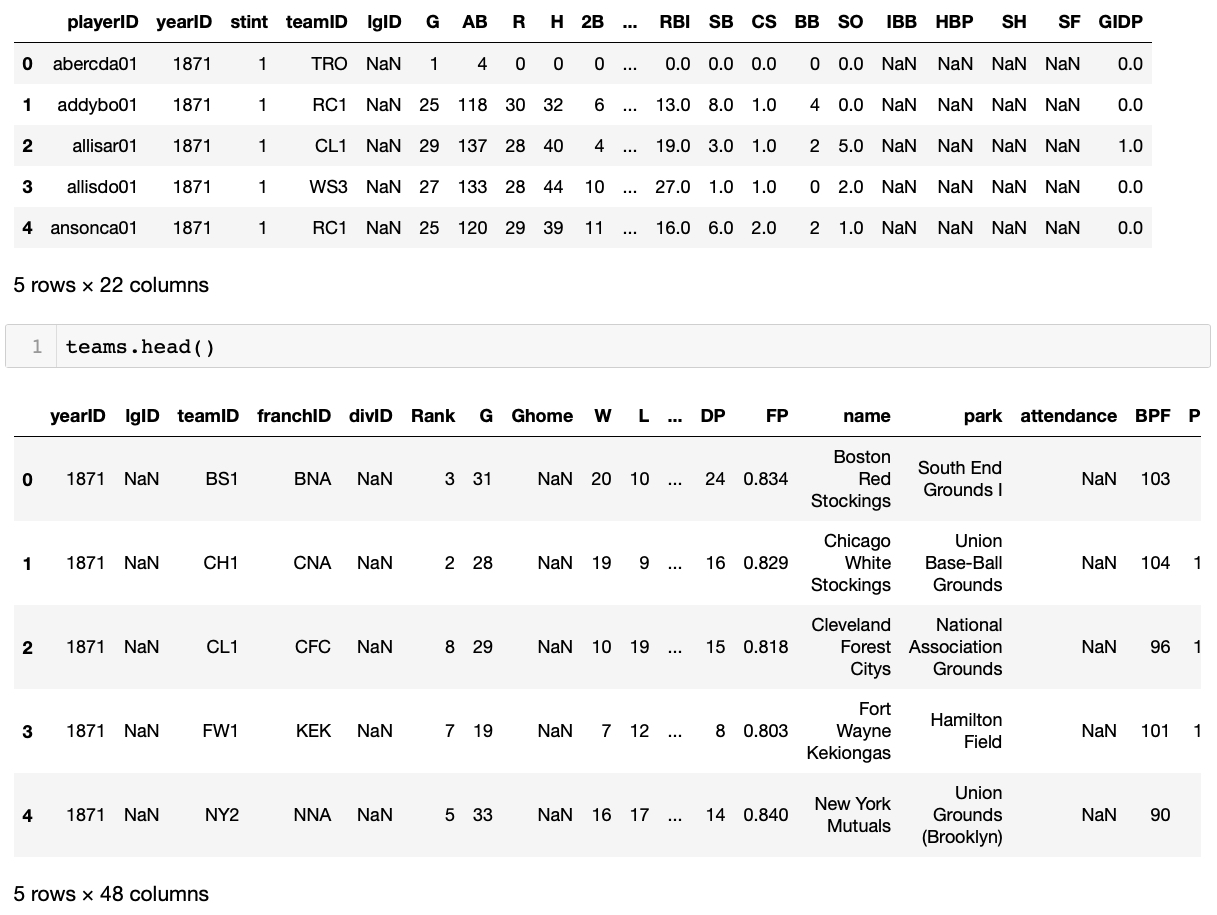
And we can get some basic summary information about these various dataframes with the info() method. We see, for example, that the table of batting data contains 107429 rows, 22 columns, and uses up approximately 18.0 MB of memory:
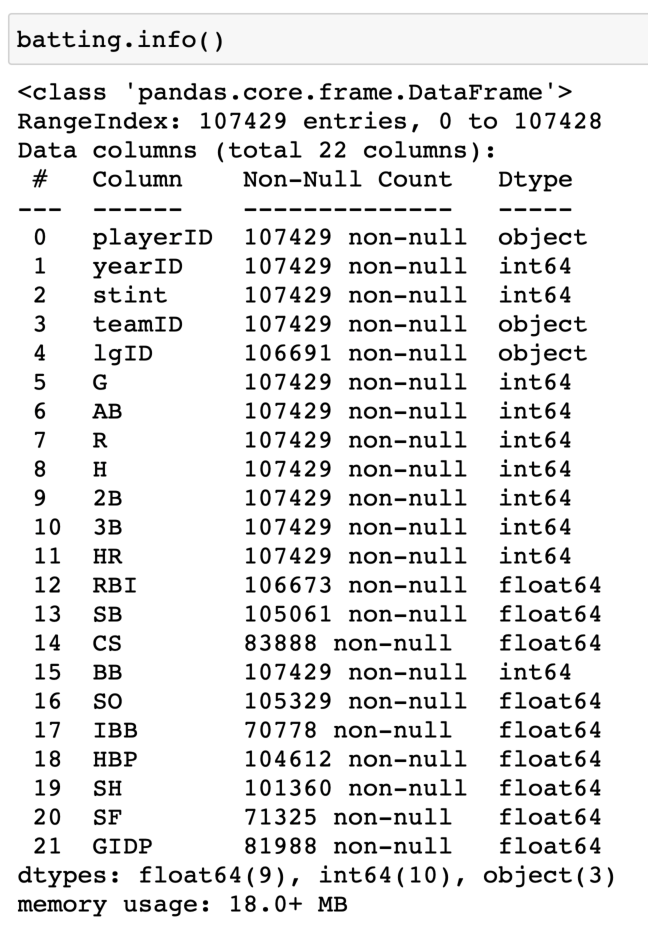
Click image for larger view (opens in new tab)
Reading Excel Spreadsheets Into Dataframes
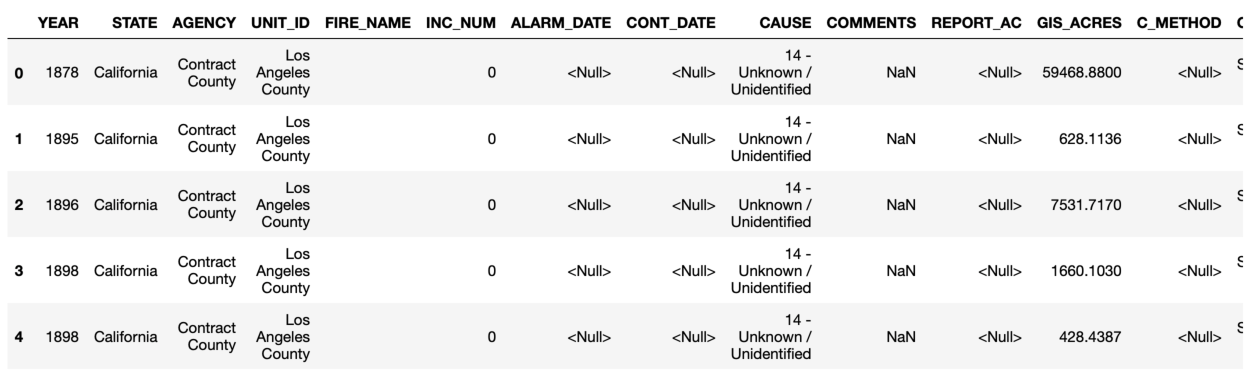
Whereas the Baseball DataBank data are stored in csv files, the California Wildfire data are in an excel spreadsheet, but we can use a similar pandas function to read it in. A spreadsheet file can contain multiple worksheets, so you usually will want to specify which sheet name(s) to read. In the case of the wildfire data, fires from years 1878 through 2016 are stored in the sheet named "2016", and fires for the year 2017 are in the sheet named "2017". For now, we will read only the data included in the 2016 sheet, using the read_excel function. We will revisit this dataset later to examine it in more detail, but for now we can take a little peak.
df16.head()
CVW material development is supported by NSF OAC awards 1854828, 2321040, 2323116 (UT Austin) and 2005506 (Indiana University)