Cleaning Data
It is often the case the data you acquire will need to be cleaned up some before you can begin working with it. (This is the less glamorous side of the field of data science, but widely recognized as a central and often cumbersome part of the process.) Here you will learn some useful tools within pandas for cleaning up data that you have imported into a dataframe, as well as some general strategies for dealing with problems in data.
The Need for Data Cleaning
We use the somewhat nonspecific term "cleaning" to refer to a variety of operations needed to get input data into a state useful for further analysis. This might include dealing with:
- Missing data
- Nonstandard data representations
- Inconsistent data
General Comments on Missing Data
When we talk about "missing" data, we are generally referring to datasets that are sufficiently regular in structure arrays, tables, dataframes) and that in principle should be completely filled with data, but are not in fact so. (As such, we are making a distinction from "irregular" data, which do not have the same implied regular structure, and which might need to be preprocessed using interpolation, kriging, smoothing or fitting in order to render them useful for further analysis.) Data can be missing from a dataset for a variety of reasons, but different software packages might be more or less accommodating of missing data. NumPy arrays and Pandas dataframes, for example, have a defined shape, so you cannot define an array or a dataframe that has different numbers of elements in each row. You could, however, fill in missing entries with a value intended to denote that the entry is missing; Pandas, for example, uses NumPy's not-a-number np.nan to reflect missing data in a series or dataframe. NumPy, on the other hand, introduces a more complicated data structure — masked arrays — that can be used to support arrays with missing entries.
To deal with missing data, there are three broad types of strategies:
- Ignore the missing data
- Drop entries that have missing data
- Fill in the missing data (or try to get it filled in at the source)
Ignoring missing data is really only a useful strategy if you understand how it will be ignored. In various aggregation operations (e.g., sum, mean, std, etc.), Pandas will simply skip over missing data as if it is not there: to compute the mean of a column, for example, Pandas computes the mean of non-missing entries in that column. Similarly, an aggregate operation over a NumPy masked array will only include those entries that are not masked.
An appropriate strategy for dropping entries with missing data really depends on what questions you want to ask. For example, later in working with wildfire data, we will want to compute the duration of a fire by calculating the number of days between when the fire was first identified (ALARM_DATE) and when it was contained (CONT_DATE). If one or both of those dates are missing, we cannot compute a duration for that fire, so it makes sense (for the purposes of getting information about fire durations) to only consider fires for which both dates are known, and drop all the rest from that analysis.
Cleaning up the Wildfire Data
While the wildfire dataset is extremely useful, we will see below that there are some glitches in data that need to be addressed before we can dive into analyses. Sometimes these sorts of issues are obvious from the outset (e.g., maybe a csv file is missing a header at the top), but often they are only discovered after you begin working with the data and notice some peculiarities. We will work through some of the issues encountered in the wildfire dataset below, and conclude this page with some broader thoughts about cleaning data.
Previously we used a pandas function to read in the wildfire data from the sheet named "2016", which was stored in an excel spreadsheet. Let's read it in and look at it.
df16 = pd.read_excel('Fires_100.xlsx', sheet_name='2016')
df16.head()
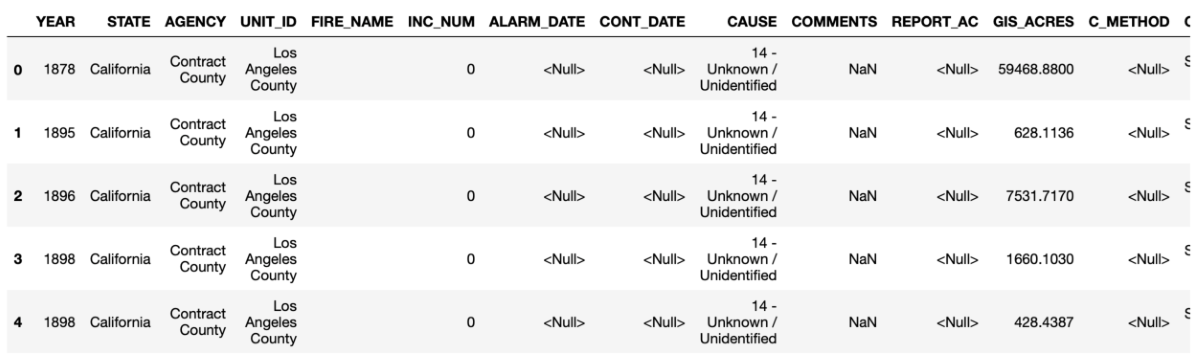
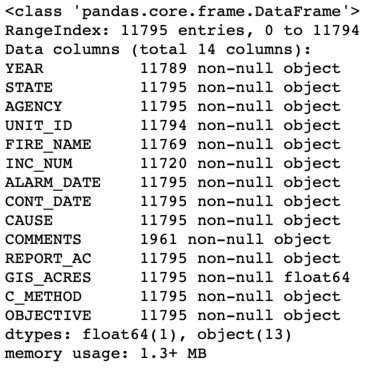
Comparing the information provided by head() and info() is useful — do you notice anything a little funny about the data? The information in df.head() suggests that there are some missing entries in the ALARM_DATE and CONT_DATE columns (encoded as <Null>), but the summary info suggests that those entries are all "non-null". Pandas can handle missing data, but it turns out that <Null> is a non-standard representation for missing data. So Pandas interprets it as the non-null character string "<Null>". Fortunately, the pd.read_excel() function has an option for specifying additional missing values. If you're following along in the Jupyter notebook, type pd.read_excel? or help(pd.read_excel) to inspect the documentation for this function. You'll see that there is an option to read_excel that allows you to specify additional data as null, missing, or na (not available) called na_values.
Now if we rerun df.head() and df.info(), we'll see now that there are many fewer instances of non-null data. Curiously, the ALARM_DATE is interpreted as an "object" (read: string), and CONT_DATE is interpreted as a date (datetype64[ns]). The reason for this discrepancy appears to be the fact that two of the ALARM_DATEs occurred before 1900, and pandas does not correctly interpret them as dates, leaving them as strings. It would be good if they were both interpreted as dates so that we could operate on them as such.
Fortunately, we can accomplish this using the pd.to_datetime function, either after we have read the dataframe in, or during the reading process itself by explicitly assigning a converter to each of the data fields:
While it was convenient to be able to do the datetime conversion directly as part of the file input, this is not always the optimal approach, as is discussed next in the section on Extracting and Reorganizing Data. As discussed on that page, if the timestamp data is in a non-standard format, you will probably want to provide a specific format string to guide the conversion rather than relying on pandas to infer a timestamp, which can take substantially more time for a large file. If you wanted to follow such an approach here, you could first call pd.read_excel to first read in the data from the file, and then do the datetime conversion for the data columns of interest by calling pd.to_datetime with the appropriate format string.
General Comments on Inconsistent Data
Data can be "inconsistent" in a variety of ways, including:
- Alternative spellings or capitalizations of text (including misspellings)
- Data in a table that are mostly of one type (e.g., floats), but with a few instances of entries with additional text included
- Data in different tables that are supposed to be linked through a set of shared keys, but there are some keys that occur in only one of the tables.
Unfortunately, there is no comprehensive solution to fixing such issues, let alone even detecting them in the first place. Clearly it is better to have such problems fixed at the data source (which might be someone or something other than you). If you do need to correct problems with data, do so in your code rather than in a data file that someone has provided for you, since any changes you make to the data file will be lost if you are provided with an updated version. We provide here a few general suggestions or strategies that might be useful in this process, however.
- If your data are in a dataframe, use
df.info()to print out the type of every column. If a unifying type cannot be inferred by pandas, column types will be generically inferred as of type "object" (read, "string"). If you think that a particular column should be of some other type (e.g., int, float, datetime64), then try to identify which entries in the column might be contributing to the lack of type coherence. - If there are alternative capitalizations of text, converting everything to a uniform state (e.g., to lower case, using a str.lower() method ) can be useful as easy form of text normalization. For more complicated issues in text normalization and/or lemmatization, various NLP packages (such as NLTK, spacy, or textblob) provide richer support.
- If your data are in a dataframe, it can be useful to examine the set of unique entries in a given column, using the
unique()method on a dataframe or series (i.e., column). This is more useful for categorical or string data than for numbers. If there are spelling variants that ought to be unified, these can be more easily spotted by filtering down to the unique set; for a dataframe df with a column named 'some_column' that you'd like to examine, the following command might be helpful:sorted(df['some_column'].unique()). By sorting the column entries, you are more likely to find spelling variants next to each other. - For more complicated variations in spelling, or for assistance in tracking down possible data entry errors, it can sometimes be useful to use string processing tools to identify closely related pieces of text. The
python-Levenshteinpackage, for example, provides a convenient and lightweight way to compute string edit distances and similarities, which can be useful in diagnosing problems with text data. - Python's built-in set objects can be useful for diagnosing inconsistencies in data. If two dataframes should have a set of matching keys (i.e., a shared set of entries in a specified column) but in fact do not, set operations can easily pinpoint discrepancies, e.g.,
set(df1['some_column'])-set(df2['some_column'])will return everything in the dataframe column in df1 that is not in the corresponding column in df2. - Plotting data is of course a time-honored method for using the pattern recognition capabilities of the human brain to quickly spot outliers or other odd looking parts of a dataset.
CVW material development is supported by NSF OAC awards 1854828, 2321040, 2323116 (UT Austin) and 2005506 (Indiana University)