Networks from Data
Datasets often directly represent, or otherwise encode, relationships among pairs of entities. A natural mathematical object, and computational data structure, for representing such relationships is a network, or graph, which consists of a set of nodes (or vertices) connected by a set of edges (or links). Being abstract and general in its definition, a network can encode many different types of entities (potentially with their own internal state or attributes) and many different types of relationships (potentially with weights, directionality, or other characteristics). The field of network science has emerged as a discipline aiming to understand the structure, function and evolution of a wide variety of networks arising in technological, biological, and sociological datasets (among others); as such, it is a field that straddles data science, mathematical modeling, and numerous domain disciplines.
There is a rich set of algorithms that have been constructed to analyze networks and graphs, including methods for:
- computing shortest paths between pairs of nodes
- identifying the set of connected components within a network
- computing various centrality measures characterizing nodes and edges (e.g., degree, closeness, betweenness, etc.)
- identifying modules or communities within networks based upon patterns of connectivity
- creating random graphs consistent with prescribed statistical properties
- much, much more
If you think there is a network lurking in your data somewhere, and you are interested in questions about network structure and properties such as those listed above, you would be well advised to use a library dedicated to the analysis of complex networks, rather than trying to code such methods on your own. Fortunately, Python is an excellent language for building and analyzing networks from data, due to its object-oriented nature, dynamic typing, support for rapid prototyping, and ecosystem of libraries. Two important libraries are NetworkX, a pure Python library, and igraph, which includes a Python wrapper around a C/C++ library. We will focus here only on NetworkX.
NetworkX is a package for the creation, manipulation, and study of the structure, dynamics, and functions of complex networks. NetworkX (conventionally shortened upon import as nx, i.e., import networkx as nx) provides a large set of functions and methods for creating and analyzing graphs, implementing methods such as those described above. A detailed description of NetworkX is beyond the scope of this tutorial, and interested readers are referred to the NetworkX documentation. Instead, we will give a very brief tour of what is possible with NetworkX, using some of our sample datasets for illustration.
Co-Player Network in Baseball
Networks encoding collaborative relationships among people have received considerable attention. The co-actor network — which connects any two actors if they were ever in a movie together — is the basis for the popular Kevin Bacon game, which identifies how many hops any actor is from Kevin Bacon. Similarly, co-authorship networks — where any two researchers are connected if they have co-authored a paper together — have been analyzed to understand the structure of scientific communities and the evolution of new fields of inquiry. With the Baseball Databank data, we can construct a similar sort of co-player network, which connects any two players if they appeared in any game for the same team during the same season. (Strictly speaking, this will slightly overestimate the number of true co-players, since two people could have been members of the same team during the same season, but not have actually been on the roster at the same time during the season or played together in a game. More fine-grained data, such as the Game Log data in Retrosheet (retrosheet.org), would be needed to resolve those sorts of associations.) Follow along in the accompanying Jupyter notebook if you would like to run the code on your own and explore the co-player network in more detail.
The basic information needed to construct this network is in the Appearances dataframe in the Baseball Databank.
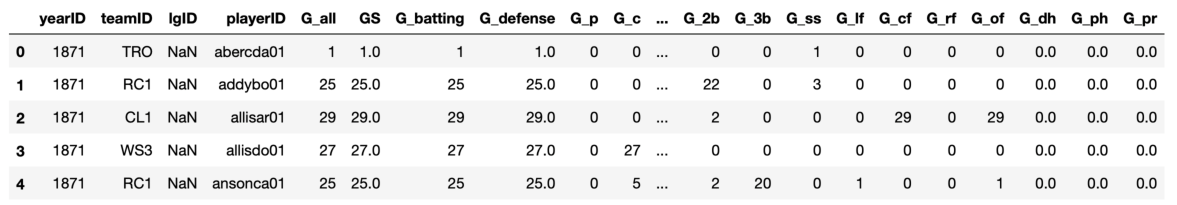
There are a couple different ways of encoding a co-player (or co-actor or co-author) network. One approach would be to have nodes that are players, with an edge between any two if they were on the same team in the same year. While conceptually straightforward, this is computationally rather inefficient, since for N players on a team during a given season, there will be N(N-1)/2 edges, such that group forms a clique in the network. Another approach is to construct a bipartite network, with some nodes representing players and other nodes representing (year, team) pairs or tuples, such that a player node is connected to a (year, team) node if the player played on the team during that year. This then replaces the N(N-1)/2 player-to-player edges with only N player-to-(year, team) edges, and also has the advantage of preserving the information about what (year, team) different players are connected through, which will be useful below.
Each row of the appearances dataframe contains information about a player (playerID) who played for a particular team (teamID) during a particular season (yearID). We can use the powerful groupby functionality in pandas to get the playerIDs of all players who were part of a given team during a given season. We can chain several methods together in one long statement, to group by year and team, select the playerIDs, identify the number of unique players for each (year, team) combination, and then reset the index to put the year and team back as columns in the new dataframe (dfyt). From this new dataframe, which can build an undirected network — represented by the networkx type nx.Graph — to connect players to (year, team) pairs, as shown below. Although the dfyt dataframe is just created as a temporary within the function definition, here is what the result looks like by carrying out the groupby operation:

Shortest paths in the co-player network
Somewhat like in the Kevin Bacon game, we can investigate the number of hops on the network required to connect any two players (although because we are including year-teams as nodes in the network, we need to adjust our counting to include only hops through the subgraph of players). For example, we can identify the shortest path (or one of possibly multiple co-shortest paths) between Ichiro Suzuki (suzukic01) and George Sisler (sislege01), the top two single-season hit leaders in MLB history:
['suzukic01',
(2001, 'SEA'),
'javiest01',
(1984, 'NYA'),
'niekrph01',
(1964, 'ML1'),
'spahnwa01',
(1942, 'BSN'),
'coonejo01',
(1928, 'BSN'),
'sislege01']
The playerIDs are a bit obscure (although well-versed baseball fans might recognize some if not all of the players on the path), but we can make things a little more user-friendly if we write a little more code to extract the players' names from the People.csv dataframe:
['Ichiro Suzuki', (2001, 'SEA'),
'Stan Javier', (1984, 'NYA'),
'Phil Niekro', (1964, 'ML1'),
'Warren Spahn', (1942, 'BSN'),
'Johnny Cooney', (1928, 'BSN'),
'George Sisler']
So we can see that Ichiro Suzuki played on the 2001 Seattle Mariners with Stan Javier, who played on the 1984 New York Yankees with Phil Niekro, who played on the 1964 Milwaukee Braves with Warren Spahn, who played on the 1942 Boston Braves with Johnny Cooney, who played on the 1928 Boston Braves with George Sisler. The acccompanying Jupyter notebook contains several other explorations using this co-player network. In addition, because year-team pairs are also nodes in the network, shortest paths can be computed between different year-team nodes. Feel free to dig around further in the notebook if you are interested.
As it turns out, Baseball-Reference.com maintains its own version of a co-player network, which is makes available through the Oracle of Baseball. Readers can enter two players into their website, and have produced a shortest path connecting those two players. The Oracle of Baseball, however, is not configured to find the shortest paths between year-teams, as is our bipartite co-player network.
Influencers in a Twitter Network
Social media are becoming a major focus of network analysis. Bots, or 'state-backed' accounts are gaining notoriety for exerting influence on social media networks such as Twitter. We will explore our Twitter dataset collected using the Twitter API to demonstrate how to characterize the network of retweets and to get some insight into influencers within the network. Interested readers might want to follow along in the accompanying Jupyter notebook to carry out these analyses or see some of the relevant code in more detail.
The tweets we are examining can be read into a pandas dataframe as shown previously in our material on interactive visualization, and we can filter that further just to get the subset of tweets that are retweets. While we can get some useful statistics just from the retweet dataframe, it is useful to package some of that data into a network for further analyses.
We can create a weighted edge list linking twitter users in our retweet dataset, where a directed edge from a source user (retweeter) to a target user (retweeted) is included if source retweets target, with additional edge information in the form of a weight that indicates how many times source retweeted target in our dataset. This can be done very succinctly using the pandas groupby functionality, allowing us to group by (source, target) pairs. The code cell below computes this edge list (in the form of a dataframe) and displays the head.
From this edge list, we can construct a directed graph (object of type nx.DiGraph) with the included edge weights, using the networkx function nx.from_pandas_edgelist. We want to create a directed graph because these retweet edges have a meaningful directionality (from retweeter to retweeted). We can also print out some basic statistics about the graph, such as the number of nodes and edges. If we do so, we find that the network has 170995 nodes and 264644 edges.
It is often of interest as to whether a network forms one complete, connected component, or whether it is broken up into multiple disjoint components. For a directed network such as the one we have constructed here, there are two different types of connnectivity to consider: weak connectivity and strong connectivity. The weakly connected components of a directed graph are comprised of those nodes that can be reached from one another by traversing edges independent of the directionality of those edges (i.e., as if we converted the directed network to an undirected one). The strongly connected components are comprised of those nodes that can only be reached from one another by following the directed edges of the graph, which is a much stronger constraint on connectivity. We can evaluate and print the number of both weakly and strongly connected components in our graph, using predefined functions in nx, as well as compute the sizes of the largest of the weak components. If we do so, we see that there are 5059 weakly connected components and 168769 strongly connected components. Diving down into the largest of the weakly connected components, we see that the network is dominated by a single large component containing 157841 nodes (or more than 92% of the full network).
5059 168769 [157841, 48, 37, 28, 26, 25, 25, 21, 20, 20]
Network Degrees
The degree of a node in a graph represents how many other nodes it is connected to. For a directed graph, there are two degrees, the in-degree and out-degree, depending on whether an edge is incoming or outgoing from a node. Twitter users with large in-degree are those who are retweeted a lot, and thus are perhaps serving as important influencers within the social network. Because our graph has weighted edges, we can compute either simply the in-degree (number of incoming edges) or the weighted in-degree (sum of all weights on incoming edges), the latter of which would indicate the total number of retweets from that user. As an exercise in the notebook, readers can examine how to compute weighted degrees.
[('UNFCCC', 9165),
('NancyPelosi', 8880),
('PaulEDawson', 5876),
('jessphoenix2018', 5312),
('MarkRuffalo', 3678),
('SenMarkey', 3100),
('RealMAGASteve', 2848),
('ed_hawkins', 2396),
('TravelWithXtina', 2175),
('SenWhitehouse', 2132)]
Closeness
Closeness centrality is a measure of the extent to which a particular node is close to other nodes. The node which requires the fewest overall 'hops' for other users to reach it has the highest 'closeness'. In order to perform a global closeness centrality calculation on an entire graph, the graph must be fully connected, which we have already demonstrated is not the case. We can still perform calculations on individual nodes. For this example, we will calculate closeness centrality for the top 10 retweeted users in our dataset.
0 PaulEDawson 0.06906497612855028
1 UNFCCC 0.11144289912839853
2 NancyPelosi 0.06334007674767712
3 jessphoenix2018 0.06303324047572335
4 MarkRuffalo 0.053675460141634726
5 SenMarkey 0.06499481526229721
6 Poseidon_NGO 0.037780729827207135
7 RealMAGASteve 0.02952882731050731
8 SenWhitehouse 0.04979530328053893
9 MikeHudema 0.06537302087006597
Network Visualization
While NetworkX is excellent for computations on graphs and networks, its focus is less tilted towards the sort of network visualization capabilities provided by some other packages. As we noted previously in Part 1 of this tutorial, network visualization is a complex problem, with many different possible approaches. Some networks are physically embedded in a two- or three-dimensional space, where nodes and edges have physical locations: a network of highways connecting different cities is one example, and the power network sending electricity between different power plants, transformers, etc. is another. Visualizing those networks is reasonably straightforward if you have information on all the positions of the nodes.
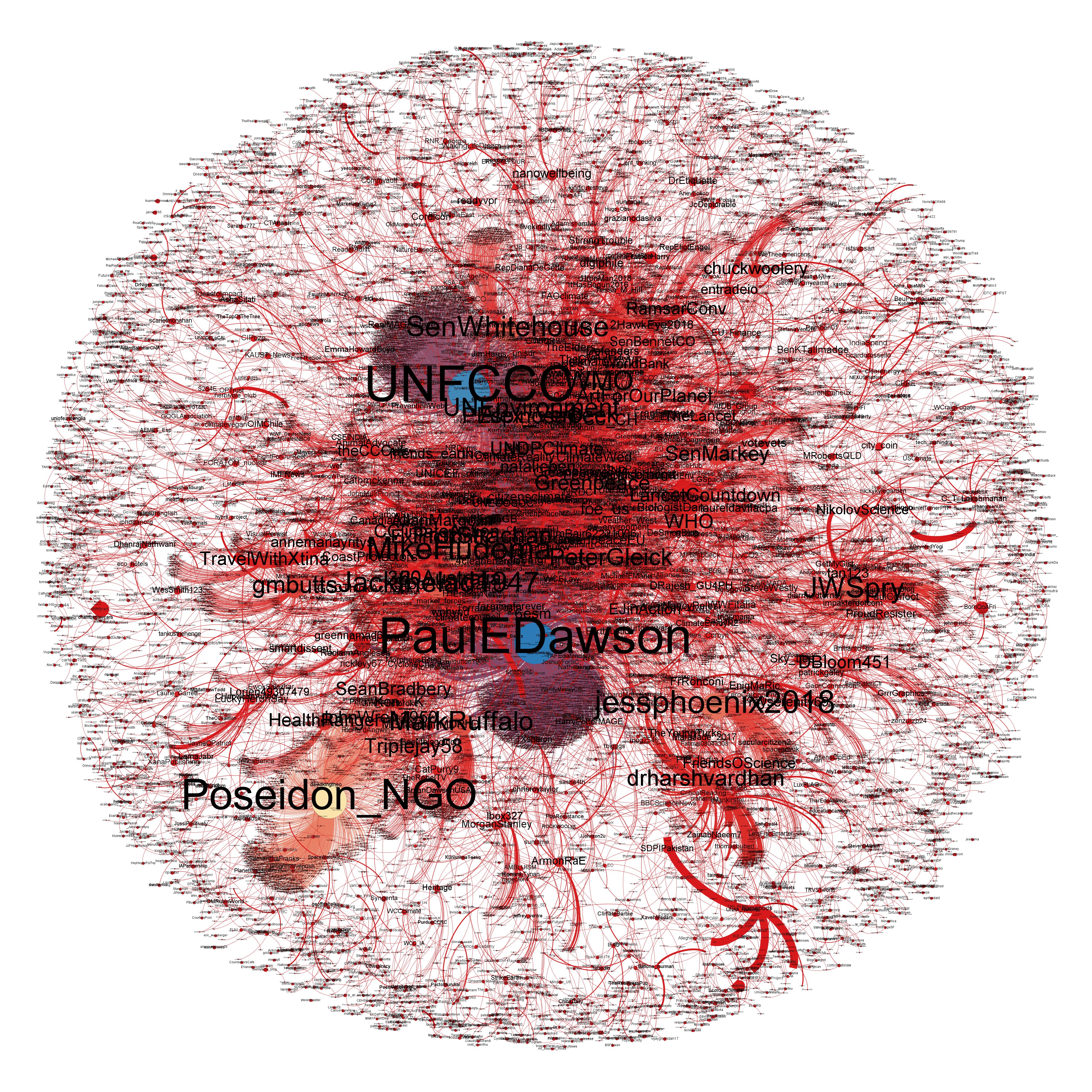
But many networks — such as the retweet network we are considering here — live in some more abstract, higher-dimensional space. Network visualization techniques that project down from a large number of dimensions to render a picture in 2 or 3 dimensions are non-unique, and can be devised to achieve a variety of different goals. Done poorly, visualization of complex networks often ends up looking like a "hairball" conveying little information. Done well, it can provide intriguing and compelling insight into the structure of a complex system. Without going into detail here, we present a visualization of our #climatechange retweet network using a different package named Gephi. Fortunately, because many different network packages are able to read and write network data in standard formats, we can write out the network data that we have assembled here in NetworkX and read it into Gephi for further visualization. The image below provides some insight into the way that some of the individuals identified as influential in the preceding analyses play a central role in the structure of the network.
CVW material development is supported by NSF OAC awards 1854828, 2321040, 2323116 (UT Austin) and 2005506 (Indiana University)